LinDA
LinDA (LINkage Disequilibrium-based Annotation) browser.
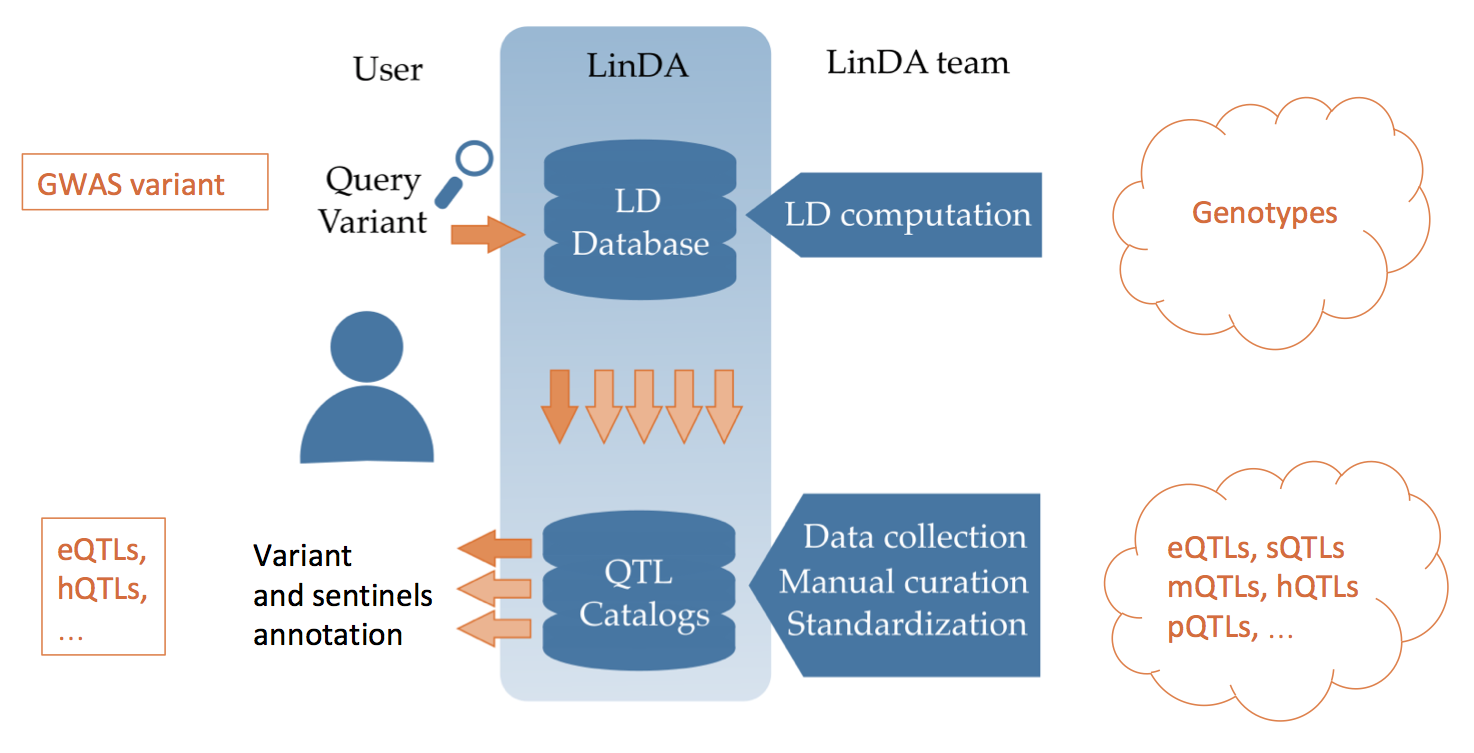
LinDA is a tool that, for a given variant, determines all variants in Linkage Disequilibrium and whether they intersect with other association studies. A large manually curated collection of QTL and association studies can be easily interrogated and results displayed with interactive plots and tables. LinDA also offers additional tools, such as computing variant frequency and pair-wise phasing, thus facilitating interpretation of results.
Tools
Overview
The LD browser is the main tool implemented in LinDA. For a given variant, it determines all the variants in Linkage Disequilibrium and whether they intersect with other association studies (QTLs and GWAS). The user can specify range of LD and the specific population in which the LD has to be retrieved. There are 32 populations of the 1000 Genomes project than can be queried for LD.
The output of the LD browser is a sequence of table each reporting the intersection with the other association studies (e.g. our eQTL Catalog). In each table are reported details about the statistical association, the gene annotation and the experiment metadata.
The eQTLs catalog
The expression Qualitative Traits Loci (eQTLs) are genetic polymorphisms associated with changes in gene expression levels and they have been successfully used to interpret the molecular role of Genome Wide Association Studies (GWAS) variants. Up to date few eQTLs databases exist but they collect only a small portion of the available datasets.
We collected several studies and here is the description of the most important features.
| Feature | Counts |
|---|---|
| Projects | 56 |
| Human populations | 15 |
| Body districts | 35 |
| Datasets | 282 |
| eGenes | 38,255 |
| eQTLs | 673,410 |
The molecular QTLs catalogs
We are continuously collecting the results of QTLs and disease association studies. A brief summary about the traits collected and the number of associations found is reported in the table on the right.
| Type of Catalog | Type of trait | Associated trait |
Associated variants |
|---|---|---|---|
| eQTLs | Whole gene RNA level | 38,255 (1) | 748,308 |
| sQTLs | Transcript level | 24,009 (1) | 188,577 |
| aseQTL | Allele-specific expression | 523 | 689 |
| polyAQTL | Alternative polyA | 2,414 | 3,820 |
| repeatQTL | Repeats expression level | 6,069 | 4,811 |
| pQTLs | protein expression level | 1,561 | 879 |
| dhsQTLs | DnaseI hyupersensitive sites | 6,069 | 6,069 |
| hQTLs | Histone modifications | 58,197 | 60,121 |
| mQTLs | DNA methylation | 219,508 | 316,376 |
(1) Results are collapsed at gene-level